Below are some recent updates, then a link to a news archive.
13/07/2025
Since my last update, I have spent a lot of time working on a new website for the Palaeontological Association (since 2020 I have been a trustee for the charity and the organisation's internet officer). We have previously run a highly customised website on our own servers, alongside a custom built webshop/membership system. This had become dated, and relied on a specialised skill set that few people in the palaeontological community possess. The new website – launched last week – addresses this by switching to an externally hosted open source framework. It was a lot of work but, I hope, places the charity on a firm digital footing for the next decade. Please do check it out by clicking on the preview below, and let us know if you spot any errors:
A new paper, to which I have contributed, has also been published. It was led by Billy Rayner, now a PhD student at UCL. In it we use high resolution synchrotron tomography to study inclusions in diamonds from the Earth's mantle:
Rayner, B.D., Kohn, S.C., Garwood, R.J., Burnham, A.D., Bulanova, G.P., Smith, C.B. & Thomson, A.R. 2025. Correlative tomography for polymineralic inclusion composition in sublithospheric diamonds. Geophysical Research Letters 52(11): e2025GL114704.
28/03/2025
A new paper led by Morena Nava, now a PhD student at Universitat de Barcelona, and with colleagues at Bristol, has just been published. In this paper we take a look at the timescale of the evolution of sea spiders, using a molecular clock and updated calibrations based on the recent work led by Romain Sabroux. Despite the changes in calibrations, however, the results are very similar to previous estimates. The group has a limited fossil record, and new discoveries may upend our understanding of these timings! Clicking on the journal name in the citation below, will take you to the open access publication.
Nava, M., Álvarez-Carretero, S., Garwood, R.J., Donoghue, P.C., Sabroux, R. & Pisani, D. 2025. A timescale for the evolutionary history of sea spiders (Arthropoda: Pycnogonida). Evolutionary Journal of the Linnean Society 4(1): kzaf001. doi: 10.1093/evolinnean/kzaf001
18/01/2025
Since COVID, I have made all my teaching materials freely available in the Learning Resources section of this website. I update as much of this as often as I am able to, and am currently making updates based on my teaching from last term. I have just posted a new website on the history of palaeontology. This site looks at human interactions with fossils before the advent of science, early scholarly thoughts on the nature of fossils, and then the development of geology and palaeontology as distinct subdisciplines. If you would like to see how this new site fits into the broader materials I provide, please do check out Learning Resources. Please explore and reuse as you see fit.
12/01/2025
A new paper led by colleagues Texas A&M University at Qatar, has just been published in the journal Artificial Intelligence Review. This contribution provides an overview of how deep learning approaches can assist with fossil image analysis, explaining the techniques used, and providing an overview of how they have been applied to date. The full citation is shown below, and you can get the open access paper by clicking on the journal name:
Yaqoob, M., Ishaq, M., Ansari, M.Y., Qaiser, Y., Hussain, R., Rabbani, H.S., Garwood, R.J. & Seers, T.D. 2025. Advancing paleontology: a survey on deep learning methodologies in fossil image analysis. Artificial Intelligence Review 58(3): 83.
Also, the simulation paper I mentioned below can now be found in an issue. If trophic tiering is your jam, please do check it out:
Furness E.N., Speed, J.D.M., Garwood R.J. & Sutton M.D. 2025. The role of evolutionary processes in determining trophic structure. Oikos 2025(1): e10597. doi: 10.1111/oik.10597
I am back in Berlin (which is currently very snowy, below). I will be working here in the the Museum für Naturkunde until September for the second half of my Alexander von Humboldt-Stiftung Research Fellowship.


09/12/2024
A new paper, led by my colleague Jason Dunlop from the Museum für Naturkunde, Berlin, has just been published in the journal PeerJ:
Dunlop, J.A. & Garwood, R.J. 2024. A review of fossil scorpion higher systematics. PeerJ 12:e18557. doi: 10.7717/peerj.18557
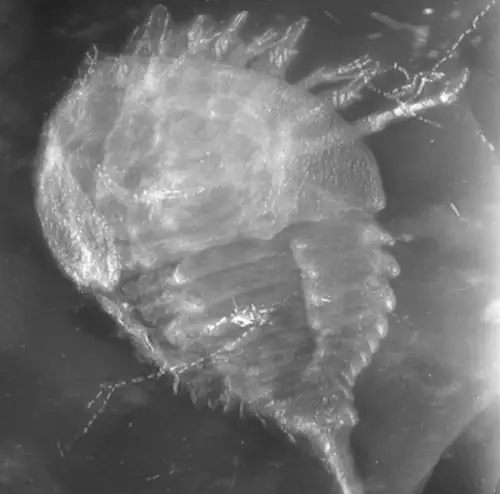
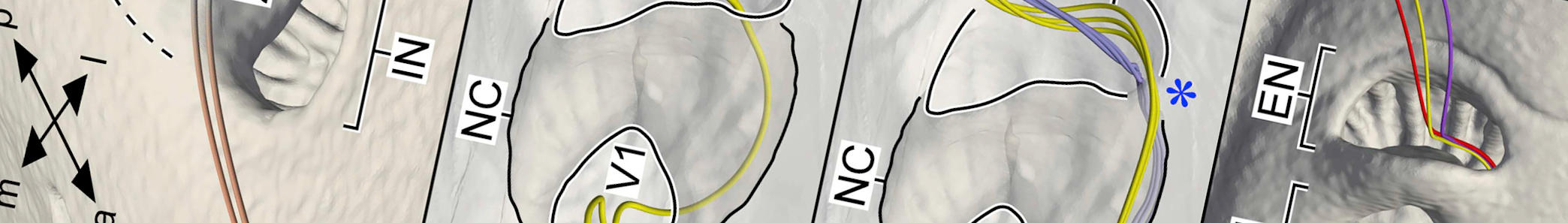
In it we provide an overview of the taxonomy of fossil scorpions. With a complicated history of study, and numerous competing schemes – some formal, others informal – the current state of the taxonomy of the group is complex and messy. We hope that by providing a history and summary of the current taxonomy alongside potentially informative characters, that this can provide a first step in clarifying the situation. It is also the first paper to arise from my Alexander von Humboldt-Stiftung Research Fellowship for experienced researchers. Clicking on the image below, which shows a series of Palaeozoic scorpion fossils from the paper, will take you to the open access publication:
14/10/2024
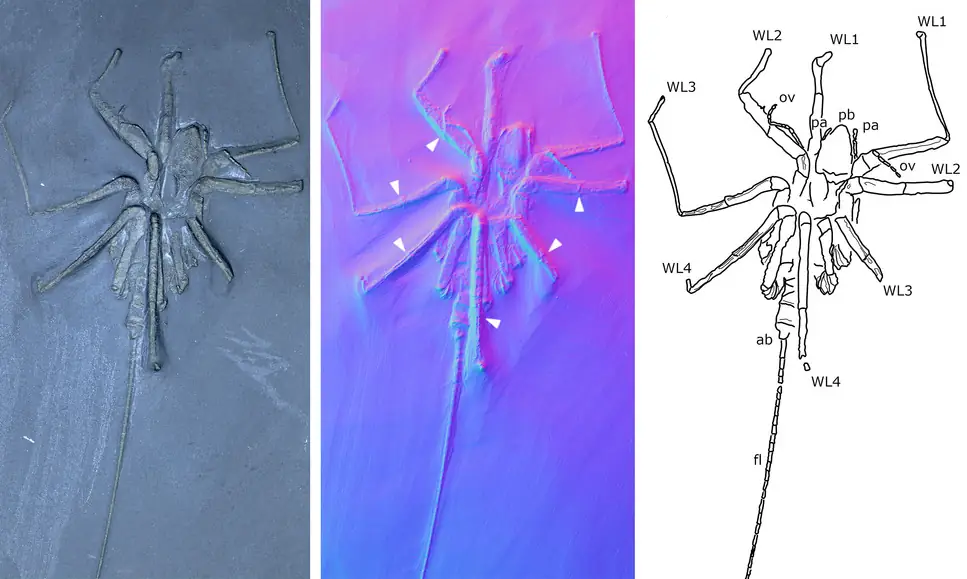
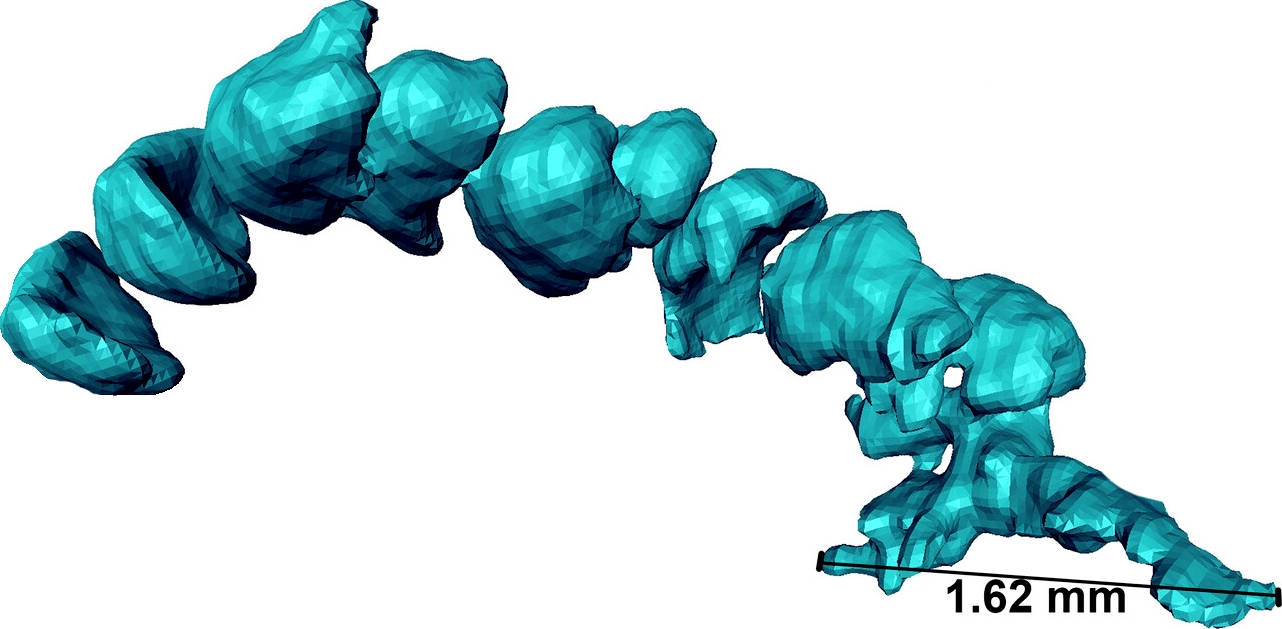
Four new papers have appeared in the last week. The first, led by Mickaël Lhéritier of Université Lyon, is on some beautiful fossils from the Montceau-Les-Mines fossil deposit in France. In particular, we use high resolution CT scanning to investigate some 305 million year old representatives of the genus Arthropleura. These animals were thought to be closely related to millipedes, and grew over two metres in length (a new fossil colleagues and I described in 2021 probably reached 2.6 metres), but for a long time we have been without a clear picture of the morphology of their head appendages, which are key to placing them on the tree of life. The μCT scans we report in this paper include some details of those, and allow us to use a total evidence analysis (one using DNA and morphology) to suggest the extinct is closely related to either just the millipede, or to a grouping of millipedes and centipedes. Details of the paper are as follows:
Lhéritier, M., Edgecombe, G.D., Garwood, R.J, Buisson, A., Gerbe, A., Mongiardino Koch, N., Vannier, J., Escarguel, G., Adrien, J., Fernandez, V., Bergeret-Medina, A. & Perrier, V. 2024. Head anatomy and phylogenomics show the Carboniferous giant Arthropleura belonged to a millipede-centipede group. Science Advances 10(41): eadp6362. doi: 10.1126/sciadv.adp6362
If you would like a more accessible overview of the paper, the lovely article linked below by our colleague James Lamsdell provides a great overview:
Lamsdell, J.C. 2024. Bring me the head of Arthropleura. Science Advances 10(41): eads9192.
In another publication, led by Euan Furness from the University of Cambridge, we use use the simulation software REvoSim to study the role of evolution in the appearance of trophic tiers. In uses a series of experiments to investigate whether supply of energy from the underlying trophic level limits biomass in each tier, or whether this is controlled by predators in the overlying trophic level. We find that which of these is the case differs between trophic levels. Full details can be found in the paper:
Furness E.N., Speed, J.D.M., Garwood R.J. & Sutton M.D. 2024. The role of evolutionary processes in determining trophic structure. Oikos. doi: 10.1111/oik.10597
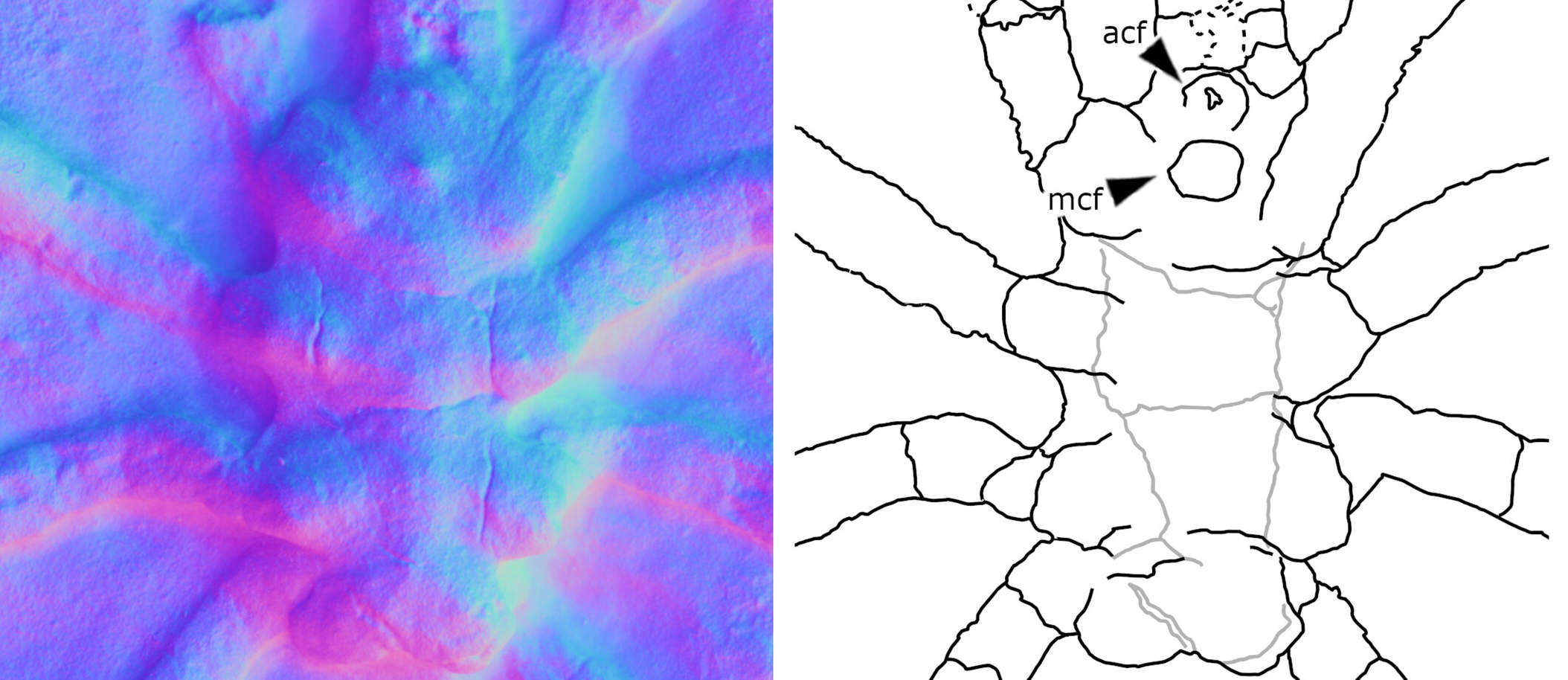
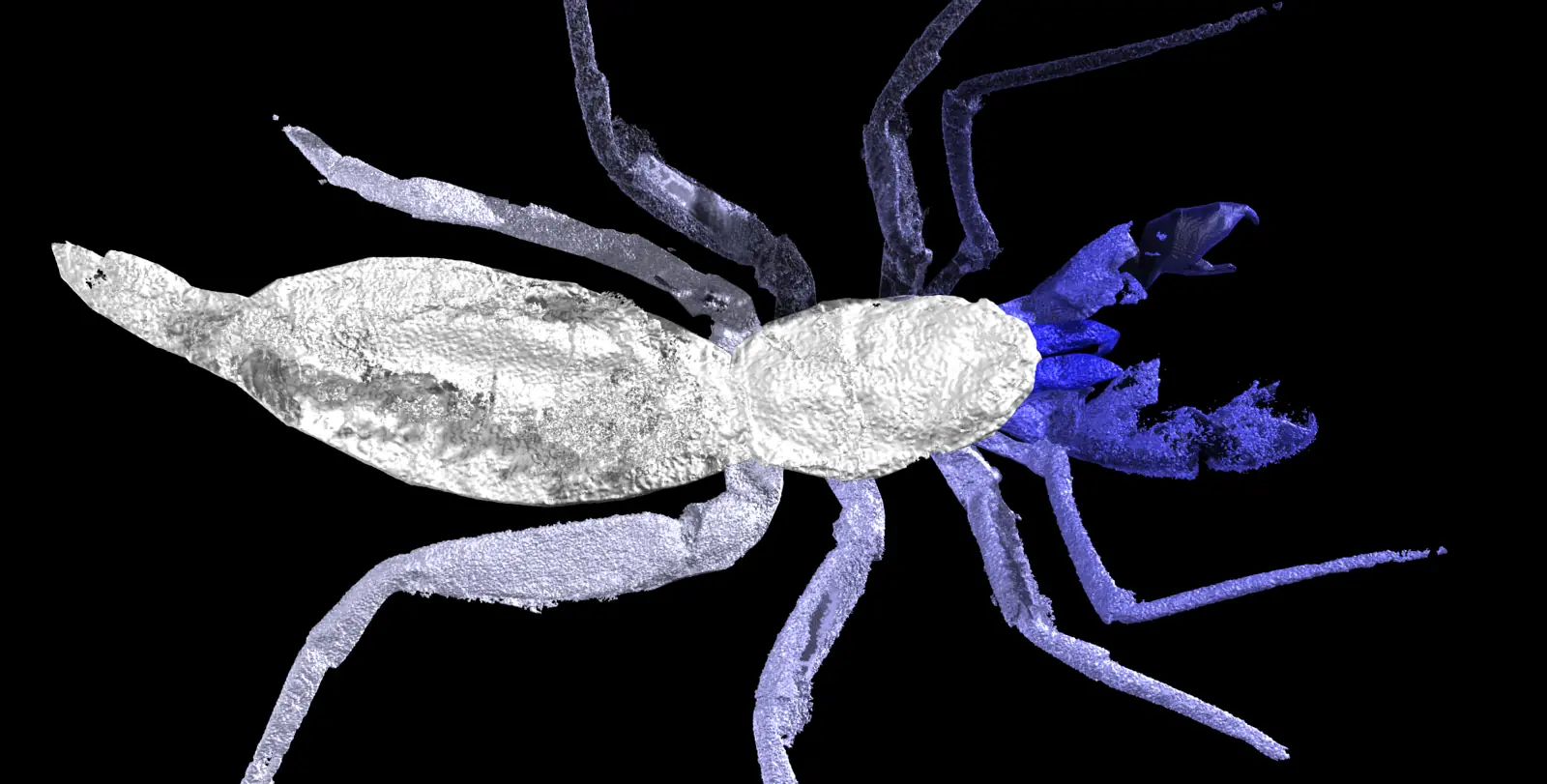
In the third paper, led by Romain Sabroux at Bristol, we reinvestigate some of the earliest sea spiders in the fossil record: those from the ~400 million year old Hunsrück Slate in Germany. We do this using μCT and Reflectance Transformation Imaging, and revisit a large number of these really wonderful fossils.
Sabroux, R., Garwood, R.J., Pisani, D. & Edgecombe, G.D. 2024. New insights into the Devonian sea spiders of the Hunsrück Slate (Arthropoda: Pycnogonida). PeerJ 12:e17766. doi: 10.7717/peerj.17766
Finally, a new paper led by my colleague Zen Goodwin at the University of Manchester, tackles the origin of organics in Martian sediments. Results based on an array of analytical techniques, suggest that these are abiotic in origin. More in the paper below.
Goodwin, A., Schröder, C., Bonsall, E. Garwood, R.J. & Tartèse, R. 2024. Abiotic Origin of Organics in the Martian Regolith. Earth and Planetary Science Letters 647:119055. doi: 10.1016/j.epsl.2024.119055
That's it for now, but there are a few more on the way.
21/09/2024
Two papers have just been published. The first – a real team effort – documents a new release of the Palaeoware package TREvoSim:
Garwood, R.J., Spencer, A.R.T., Bates, C.T.M., Callender-Crowe, L.M., Dunn, F.S., Halliday, T.J.D., Keating, J.N., Mongiardino Koch, N., Parry, L.A., Sansom, R.S., Smith, T.J., Sutton, M.D. & Vanteghem, T. 2024. TREvoSim v3: An individual based simulation tool for generating trees and morphological data. Journal Of Open Source Software 9(101): 6722. doi: 10.21105/joss.06722.
This release includes a host of new features. These include: tools for studying evolutionary processes and the nature of evolution; options to provide more user control over the simulations and their outputs; a fully updated suite of tests covering all aspects of the simulation, and expanded documentation; and a significantly improved codebase courtesy of the detailed input from the reviewers of the above publication. The paper is open access, and the new release of the software can be accessed by clicking the image below.
The second paper, led by Tom Smith at the University of Oxford, uses a new addition to TREvoSim to study the evolutionary outcomes of ecosystem engineering – i.e. organism environment feedback. Through a series of experiments, we suggest that there is no general expected outcome when ecosystem engineers are introduced to an ecosystem. In some replicates, ecosystem engineers dominate, in others they merely survive, and in many they just die out. Clicking on the journal name in the citation below will take you to the open access paper.
Smith, T.J., Parry, L.A., Dunn, F.S. & Garwood, R.J. 2024. Exploring the macroevolutionary impact of ecosystem engineers using an individual-based eco-evolutionary simulation. Palaeontology 67(5): e12701. doi: 10.1111/pala.12701
04/07/2024
Another paper, led by my colleague Zen Goodwin at the University of Manchester on a ~1200 million year old impact deposit found in Northern Scotland – the Stac Fada Member – was just published. In it, we use the geochemistry of this rock to suggest the nature of the impactor that might have caused it. You can get more information, and read the open access paper, by clicking on the journal name in the reference below:
Goodwin, A., Garwood, R.J. & Tartèse, R. 2024. Geochemical evidence for a chondritic impactor in altered impact glass from the Stac Fada Member impactite, NW Scotland. Journal of the Geological Society 181(5): jgs2023-213. doi: 10.1144/jgs2023-213.
09/05/2024
From May – September 2024 (and then January – September 2025), I am based at the Museum für Naturkunde, Berlin, working with Jason Dunlop and Lauren Sumner-Rooney on the fossil record and phylogeny of the arachnids, and the evolution of their visual systems. I'm incredibly grateful to the Alexander von Humboldt-Stiftung for funding this visit through a Humboldt Research Fellowship for experienced researchers. It's a really exciting opportunity, and I am thrilled to be here!
Since my last update, a paper on the kinematics of amblypygid hunting, led by Manchester Metropolitan colleague Callum McLean has been published. Details below.
McLean, C., Brassey, C., Seiter, M., Garwood, R.J. & Gardiner, J.D. 2024. The kinematics of amblypygid (Arachnida) pedipalps during predation: Extreme elongation in raptorial appendages does not result in a proportionate increase in reach and closing speed. The Journal of Experimental Biology 227(4): jeb246654. doi: 10.1242/jeb.246654
08/01/2024
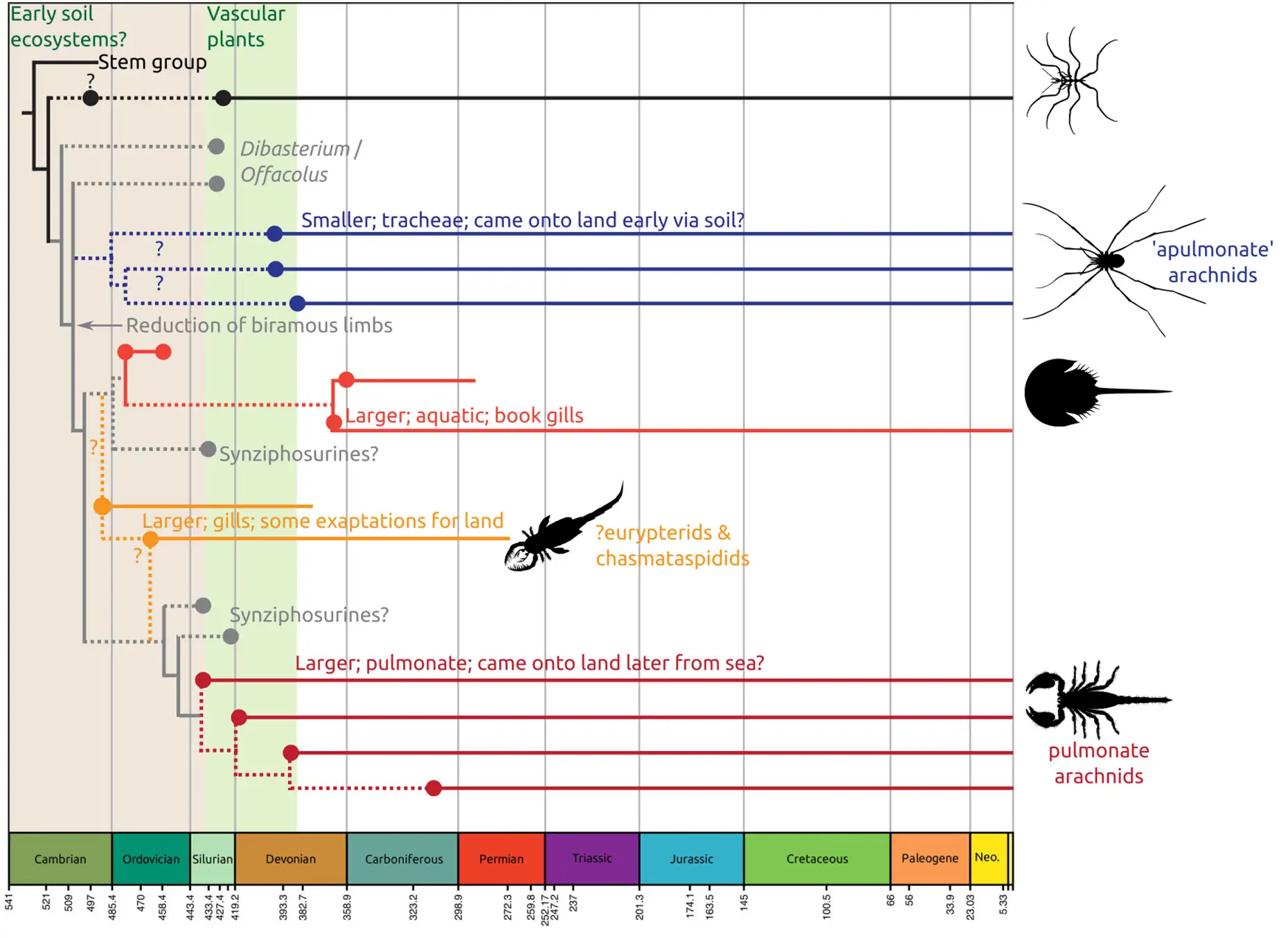
A new paper appeared at the end of last year, looking at the current status of our understanding of chelicerate phylogeny, and terrestrialisation in the arachnids (the two are linked) in the context of the fossil record. In it, my colleague Jason Dunlop and I, consider:
- The first appearance of major groups in comparison to the arachnid tree of life
- The nature of the early plant fossil record and how arachnids may have moved onto land in early terrestrial ecosystems
- The impact of including horseshoe crabs within the arachnids in this light, and what a horseshoe crab is
We suggest ultimately that recent phylogenetic results are best reconciled with fossils through inferring multiple terrestrialization events, which might have involved quite different approaches to breathing air. Click on the image below – which shows a potential scenario for arachnid terrestrialisation(s) – for a copy of the open access paper.
We suggest ultimately that recent phylogenetic results are best reconciled with fossils through inferring multiple terrestrialization events, which might have involved quite different approaches to breathing air. Click on the image below for a copy of the open access paper.
The full citation for the paper is:
Garwood, R.J. & Dunlop, J.A. 2023. Consensus and conflict between chelicerate phylogeny and their current fossil record. Arachnologische Mitteilungen 66: 2-16. doi: 10.30963/aramit6602
28/11/2023
Several new papers have been published since my last update – this has been a year of finishing off ongoing projects! I've listed these with a short overview below, in alphabetical order by first author.
In a paper in Arachnology, my colleague Jason Dunlop and I look at the taxonomy of the fossil scorpion genus Palaeophonus:
Dunlop, J.A. & Garwood, R.J. 2023. The status of two fossils assigned to the scorpion genus Palaeophonus and its interpretation as a senior synonym of Allopalaeophonus. Arachnology 19(6):940-943. doi: 10.13156/arac.2023.19.6.940
A paper led by my colleague Arthur Goodwin at the University of Manchester uses X-ray microtomography to try and better understand the origin of a martian regolith meteorite:
Goodwin, A., Tartèse, R, Garwood, R.J. & Almeida, N.V. 2023. A 3D Petrofabric Examination of Martian Breccia NWA 11220 via X-ray Computed Microtomography: Evidence for an Impact Lithology. Journal of Geophysical Research: Planets 128(11): e2023JE007916. doi: 10.1029/2023JE007916
A contribution led by Lorenzo Lustri of the University of Lausanne studies the developmental changes in Xiphosura (horseshoe crabs) throughout their evolutionary history:
Lustri, L., Antcliffe, J.B., Saleh, F., Haug, C., Laibl, L., Garwood, R. J., Haug, J. & Daley, A.C. 2023. New perspectives on the evolutionary history of xiphosuran development through comparison with other fossils euchelicerates. Frontiers in Earth Sciences 11:1270429. doi: 10.3389/fevo.2023.1270429
The preprint introduced below (14/09/2022) has now been accepted and appeared in the journal Palaeontology:
Mongiardino Koch, N., Garwood, R.J. & Parry, L.A. 2023. Inaccurate fossil placement does not compromise tip-dated divergence times. Palaeontology 66 (6): e12680. doi: 10.1111/pala.12680
And in a study led by colleagues from the University of Bristol, we interpret structures from the late Ediacaran Khatyspyt Formation of Arctic Siberia, shown below, to be body fossils (as opposed to traces):
Psarras, C., Donoghue, P.C., Garwood, R.J., Grazhdankin, D.V., Parry, L.A., Rogov, V.I. & Liu, A.G. 2023. Three-dimensional reconstruction, taphonomic and petrological data suggest that the oldest record of bioturbation is a body fossil coquina. Papers in Palaeontology 9(6):e1531. doi: 10.1002/spp2.1531
15/09/2023
A new version of the individual-based eco-evolutionary simulation software that colleagues and I have been developing for the last decade is now available, and has been published in the Journal of Open Source Software:
Furness, E.N., Garwood, R.J. & Sutton, M.D. 2023. REvoSim v3: A fast evolutionary simulation tool with ecological processes. Journal of Open Source Software 8(89): 5284. doi: 10.21105/joss.05284
It has a host of new features, including:
- A new genome architecture allowing user-defined genome lengths
- The concept of systems – processes that can be applied to specified parts of the genome
- Pathogens – a system which allows pathogen or predator/prey dynamics within experiments
- The capability to use images to spatially control simulation variables (linkages)
- Ecological interactions between organisms
- Rewritten logging and species identification code
The JOSS paper describes these additions in full, and they are also explained in the REvoSim documentation. Click on the image below to visit the paper.
27/07/2023
A new paper led by a team at Bristol on fossil sea spiders has just been published:
Sabroux, R., Edgecombe, G.D., Pisani, D. & Garwood, R.J. 2023. New insights into the sea spider fauna (Arthropoda: Pycnogonida) of La Voulte-sur-Rhône, France (Jurassic: Callovian). Papers in Palaeontology 9(4): e1515. doi: 10.1002/spp2.1515
Sea spiders are wonderfully weird aquatic chelicerates (i.e. they are more closely related to horseshoe crabs and arachnids than they are other arthropods). This work is led by Marie Skłodowska-Curie fellow in Bristol, Romain, who is reassessing the group's fossil record. In this paper, we tackle some ~163 million year old fossils from the La Voulte-sur-Rhône Lagerstätte in France. We analyse the fossils using x-ray microtomography and reflectance transformation imaging, making some changes to their taxonomy that have implications on how we use the fossils as calibrations in molecular clock analyses. Click on the picture below, taken from the paper, to read the open access publication:
10/07/2023
Two papers towards which I have contributed have just appeared. One was led by my colleague Joe Keating, now at Bristol:
Keating, J.N., Garwood, R.J. & Sansom, R.S. 2023. Phylogenetic congruence, conflict and consilience between molecular and morphological data. BMC Ecology and Evolution 23(1): 30. doi: 10.1186/s12862-023-02131-z
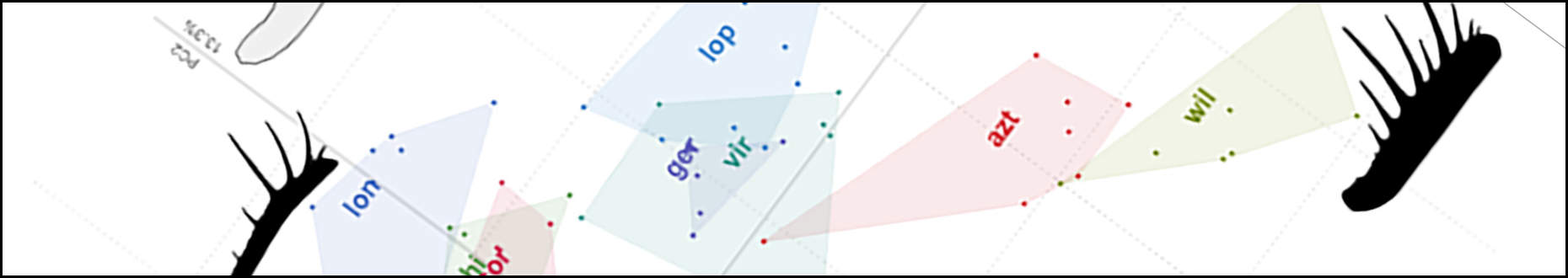
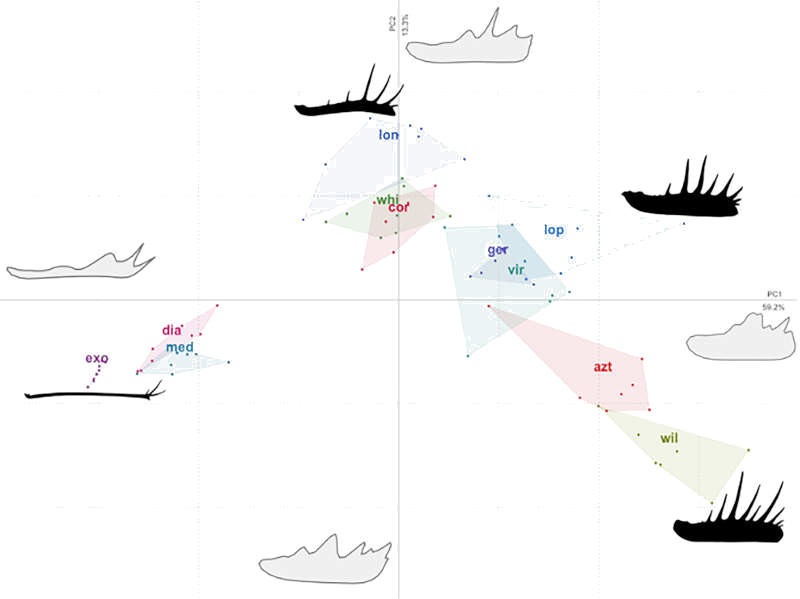
We use this paper as an opportunity to explore the congruence of molecular and morphological data in combined phylogenetic analyses using a meta-analysis of 32 datasets. We ask questions including whether morphology and molecules should be combined, how these forms of data interact when they are, and whether the results are reliable. The answers to those questions – and more – can be found in the open access paper, linked from the pretty tree-space graphs below:
The other paper, also open access, was led by my colleague Arthur Goodwin at the University of Manchester:
Goodwin, A., Tartèse, R., Garwood, R.J., Jerrett, R. & Joy, K.H. 2023. Provenance of altered carbon phases and impact history of the Stac Fada Member, NW Scotland. Meteoritics and Planetary Science.
This contribution looks at a ~1200 million year old rock deposited by an ancient impact, found today in NW Scotland, in particular the chemistry of Carbon found within the rock. Read on if organic geochemistry, and ancient rocks, are your bag!
25/04/2023
A paper that has been in the works for a rather long time has just been published. In it, my colleague Jason Dunlop and I restudy two Carboniferous whip scorpions using X-ray microtomography, and include these species in an updated arachnid phylogeny. We suggest that this group – an order called the Thelyphonida – exhibit a high degree of stasis, having changed relatively little in the last 315 million years. Click on the image below to check out the paper.
Garwood, R.J. & Dunlop, J.A. 2023. X-ray microtomography of the late Carboniferous whip scorpions (Arachnida, Thelyphonida) Geralinura britannica and Proschizomus petrunkevitchi. Journal of Systematic Palaeontology 21(1): 2180450. doi: 10.1080/14772019.2023.2180450
16/01/2023
A new paper came out over the Christmas break, led by Euan Furness, from Imperial College. This introduces a new biogeographic species-area model that partitions heterogeneity of an environment from its area. More details in the publication, linked below!
Furness, E.N., Saupe, E.E., Garwood, R.J., Mannion, P.D. & Sutton, M.D. 2023. The jigsaw model: A biogeographic model that partitions habitat heterogeneity from area. Frontiers of Biogeography 15.1:e58477. doi: 10.21425/F5FBG58477
14/09/2022
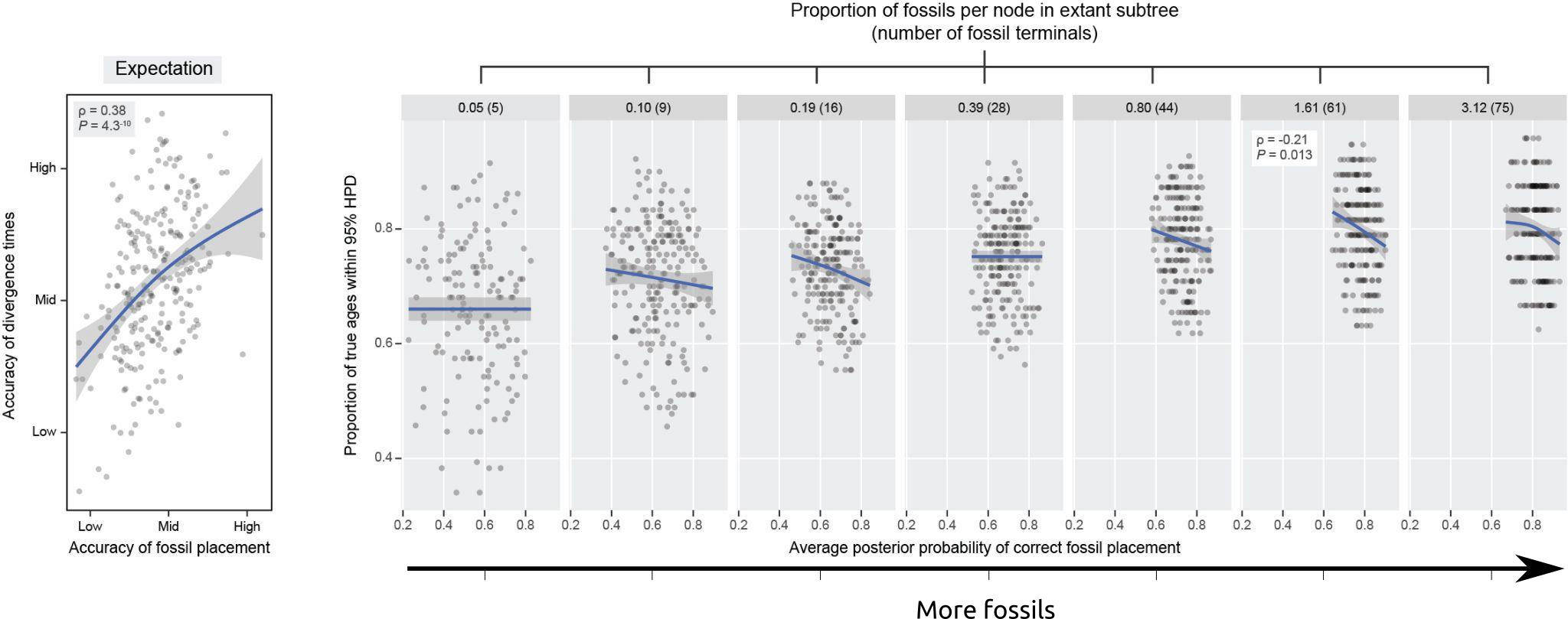
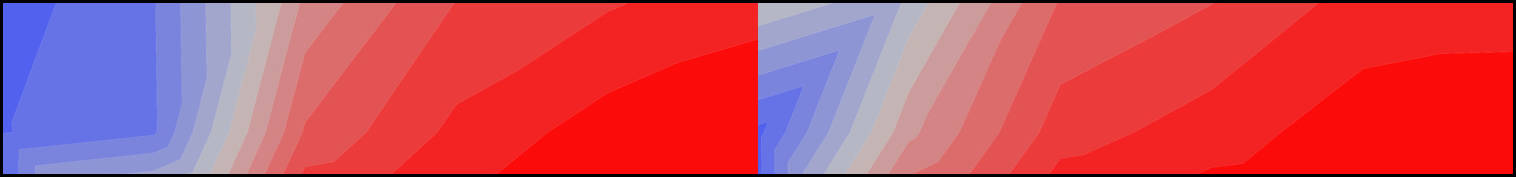
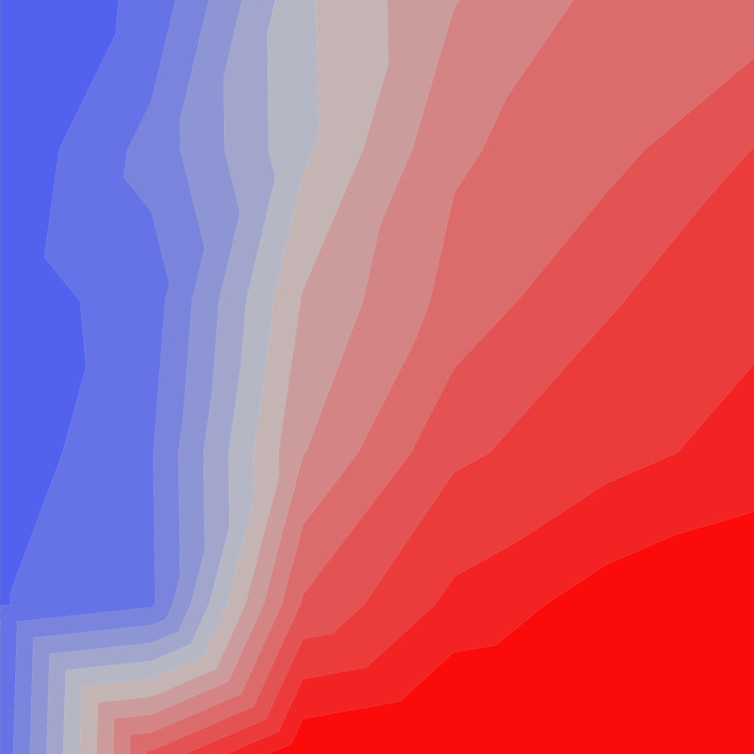
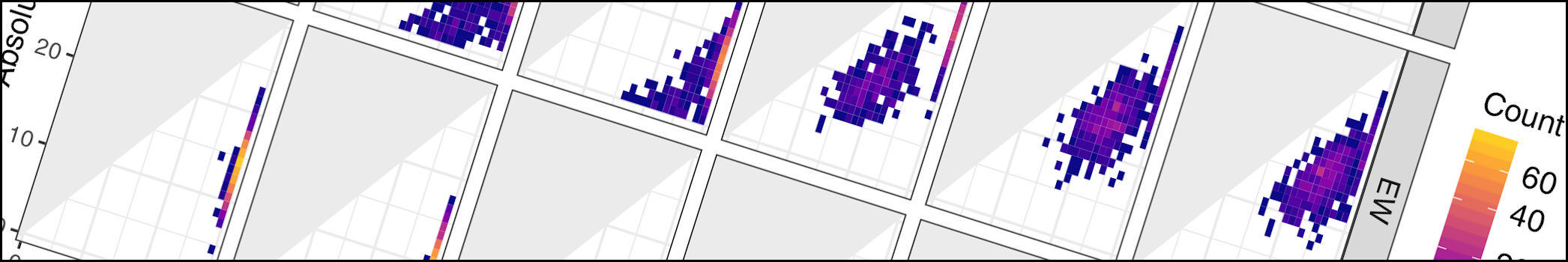
A new study with my colleagues Nicolás Mongiardino Koch and Luke Parry has just been posted as a preprint. In this contribution, we use the simulation package TREvoSim to generate phylogenetic data, and then use this to assess the impact that inaccurate fossil placement has on divergence time estimates in clock analyses. The take home message is that with empirically realistic datasets it appears that fossils are generally reliably placed, and the level of error we see does not strongly impact on accuracy of divergence time estimates (compare the expectation, and the result graphs shown below) – although we do see some interesting systematic biases. Hopefully this will be out as a paper soon.
Mongiardino Koch, N., Garwood, R.J. & Parry, L.A. 2022. Inaccurate fossil placement does not compromise tip-dated divergence times. bioRxiv. doi: 10.1101/2022.08.25.505200
10/03/2022
A new paper has appeared, led by a PhD student Arthur Goodwin. This contribution is a review of the “Black Beauty” Martian regolith breccia, providing an overview of its geology with a focus on what is says about the habitability of Mars. It's available open access:
Goodwin, A., Garwood, R.J. & Tartèse, R. 2022. A Review of the “Black Beauty” Martian Regolith Breccia and Its Martian Habitability Record. Astrobiology. doi:10. 1089/ast.2021.0069
22/12/2021
Over the summer, I was lucky enough to work with a team led by Neil Davies, from the University of Cambridge, on describing the largest known arthropod fossil to date – a Carboniferous millipede. The paper has just been published:
Davies, N.S., Garwood, R.J., McMahon, W.J., Schneider, J.W. & Shillito, A.P. 2021. The largest arthropod in Earth history: insights from newly discovered Arthropleura remains (Serpukhovian Stainmore Formation, Northumberland, England). Journal of the Geological Society. doi: 10.1144/jgs2021-115 (More information: 1, 2, 3, 4)
We estimate that this creature was about 2.6 metres in length when alive, and weighed in the region of 50 kilos. The fossil suggests gigantism in this group began before high oxygen concentrations built up in the Late Carboniferous, and we also took the opportunity to chart the changes in palaeogeography of the iconic trace fossils associated with these animals. The Natural History Museum have posted an excellent overview of the findings, and you can find further press coverage by following the links at the end of the citation above.
Below you can see an image of the fossil itself, then a reconstruction of the animal in life. Clicking on either will take you to a copy of the paper.
08/12/2021
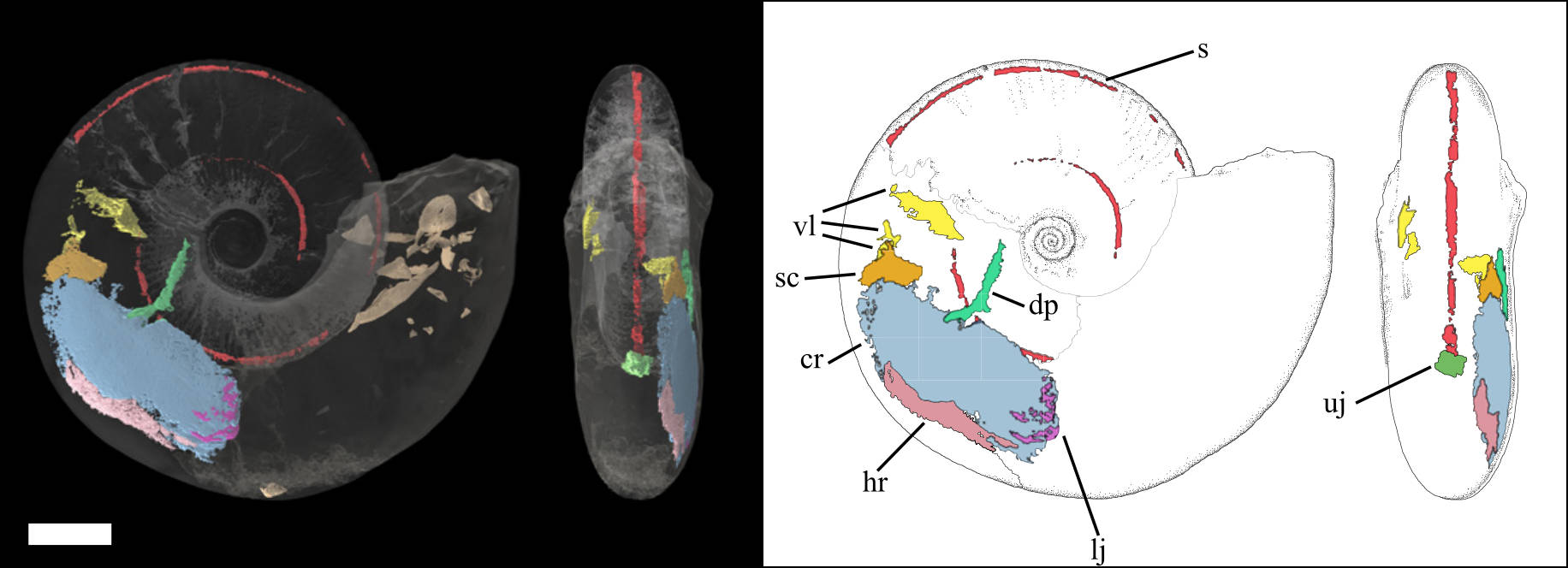
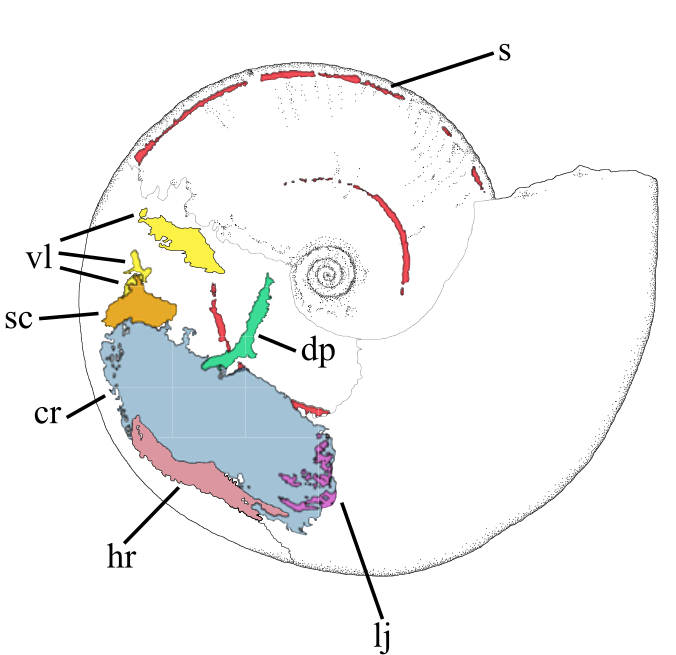
I'm very pleased to report on a new paper, led by a colleague at the University of Cardiff, Dr Lesley Cherns, that has been in the works for several years. A team of use from across the UK used a combination of X-ray and neutron tomography to scan a fossil ammonite with soft tissues preserved. This is amongst the first fossils to show the preservation of soft parts in an ammonite, and the configuration of the muscles we found in our scan tells us something about the evolution of this iconic group. For example, we suggest that the animals had a mobile, retractable body, and swam using jet propulsion: sa mechanism that must have evolved relatively early in the evolution of ammonoids and their closest relatives – the octopodes, squid and cuttlefish. The paper details are as follows:
Cherns, L., Spencer, A.R., Rahman, I., Garwood, R.J., Reedman, C., Burca, G., Turner, M., Hollingworth, N.T. & Hilton, J.M. 2021. Correlative tomography of exceptionally preserved Jurassic ammonite implies hyponome-propelled swimming. Geology. doi: 10.1130/G49551.1 (More information: 1, 2, 3)
You can learn more about the fossil by following the links at the end of the above citation, which will take you to press coverage of the discovery.
06/11/2021
A funded PhD opportunity is available for a project supervised by myself and my colleague Rob Sansom, titled The timing and modes of macroevolutionary change: molecules, morphology, and simulations. If you're interested in real-world data, simulations and evolution, please don't hesitate to get in touch if you have any questions! Deadline for applications is the 21st of January. here for more details of this PhD opportunity.
17/07/2021
A new paper, led by my colleague Callum McLean at Manchester Metropolitan University, has recently been published. The study investigates shape complexity in the pedipalps (mouthparts) of whip spiders within and across species using elliptical Fourier analysis. Pedipalps are used for hunting, but also used in contest for mates. The paper has several implications, but a key finding is that there may be a trade-off between investment in pedipalp length (for use in selection/contest) v.s. spine length (for use in prey capture). You can read more about it in the (open access) paper, which is linked below:
McLean, C., Garwood, R.J. and Brassey, C. 2021. Assessing the patterns and drivers of shape complexity in the amblypygid pedipalp using elliptical Fourier analysis. Ecology and Evolution. doi: 10.1002/ece3.7882
17/06/2021
A new paper, led by Euan Furness, from Imperial, has just been published:
Furness, E.N., Garwood, R.J., Mannion, P. D. & Sutton, M.D. 2021. Productivity, niche availability, species richness and extinction risk: Untangling relationships using individual-based simulations. Ecology and Evolution doi: 10.1002/ece3.7730
This contribution uses the Palaeoware package REvoSim to investigate the impact of productivity on species richness. It shows that species richness scales with productivity – matching some of the predictions of an explanation for this pattern called the more individuals hypothesis. For example, rare species are more extinction prone, as MIH predicts. But, we show that species richness only scales with productivity when species can partition niche space – the relationship doesn't hold when niche partitioning is prevented through saturation. Thus even when neutral explanations do help us understand patterns, we suggest niche theory is also important to consider.
04/05/2021
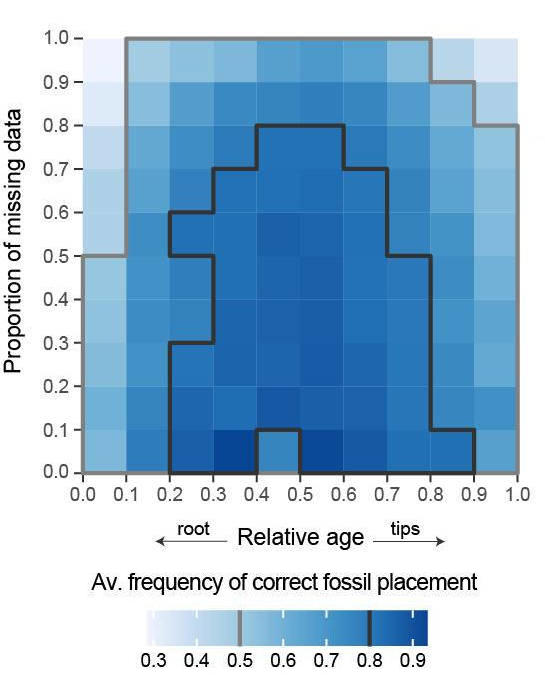
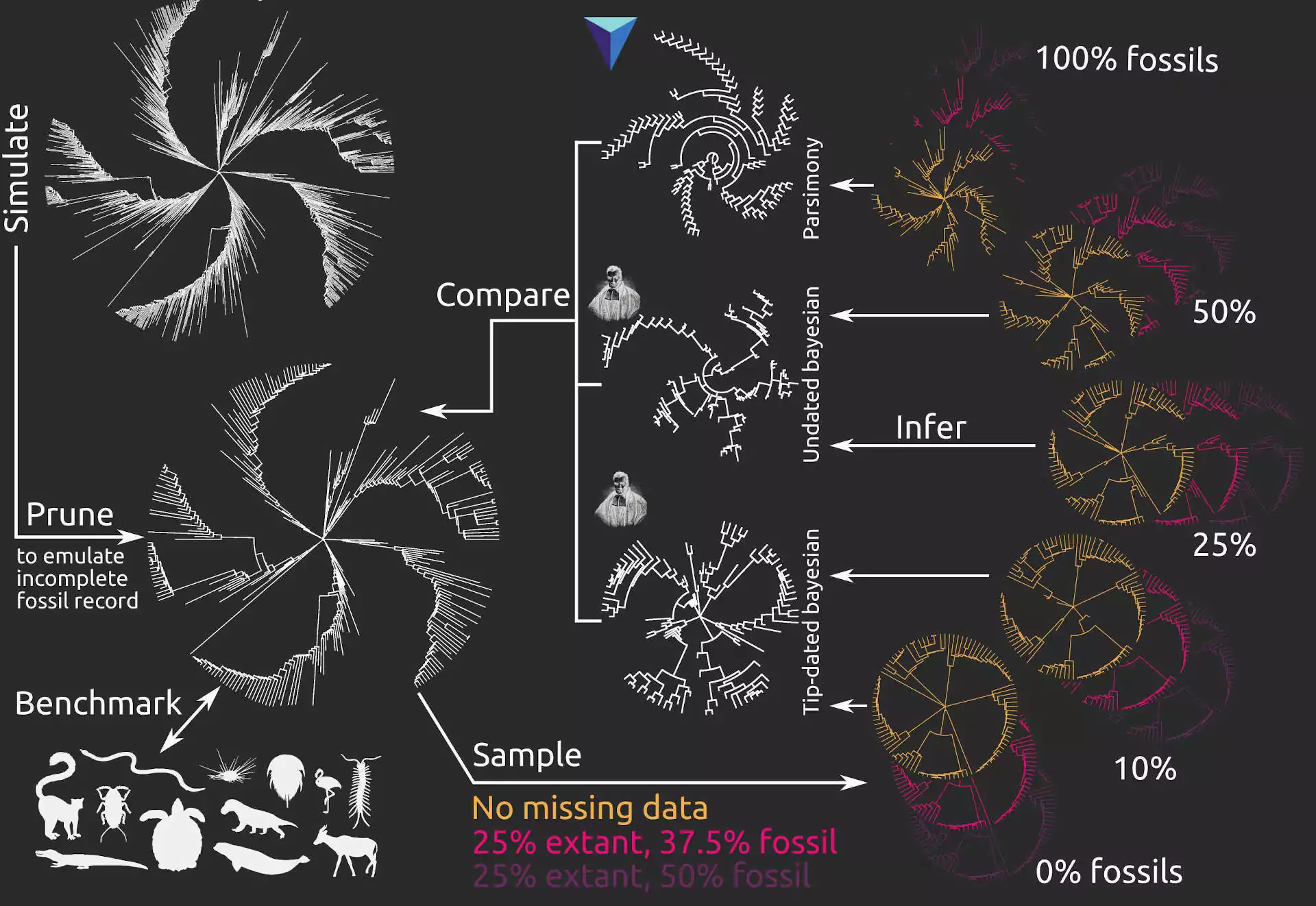
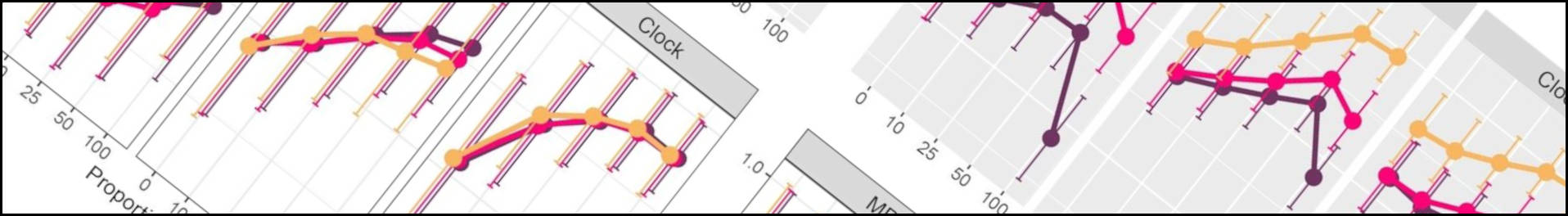
One of the preprints mentioned below has just been published in Proceedings B. Whilst fossils are key to understanding past life, their impact on phylogenetic reconstruction is hard to assess: in real organisms we lack a true evolutionary tree. In this study, led by my colleague Nicolás Mongiardino Koch, we use the Palaeoware package TREvoSim to generate trees and associated character data, benchmark these against empirical datasets, and then evaluate the impact of including fossils in phylogenies. We do so using an analytical pipeline summarised in the graphic below:
The outcome? We show that fossils – irrespective of the levels of missing data or inference method – improve the accuracy of phylogenetic inference, and induce the collapse of highly uncertain relationships that tend to be incorrectly resolved. In our study, tip-dated analyses outperform undated inference methods: we suggest that the age of fossils contains important information that aids phylogenetic analyses. You can learn more in the open access paper linked below:
Mongiardino Koch, N., Garwood, R.J. & Parry, L.A. 2021. Fossils improve phylogenetic analyses of morphological characters. Proceedings of the Royal Society B: 288(1950):20210044. doi: 10.1098/rspb.2021.0044
23/04/2021
A few new pieces of work that I've been lucky enough to contribute to have appeared since I last updated this website. The paper linked below, led by Imperial PhD student Euan Furness, uses the Palaeoware package REvoSim to investigate the impact of environmental disturbance on species richness. It shows that a key consideration is the scale of a disturbance, and the spatial heterogeneity of the environment. In a heterogeneous environment, small scale disturbance leads to a decrease in species richness, but in a homogeneous one, it increases species richness. In contrast, large scale disturbances always reduce the number of species. You can read more in the open access paper below:
Furness E.N., Garwood R.J., Mannion P.D. & Sutton M.D. 2021. Evolutionary simulations clarify and reconcile biodiversity-disturbance models. Proceedings of the Royal Society B: 288(1949):20210240. doi: 10.1098/rspb.2021.0240
A paper on smell in the sea catfish – for which I assisted with data collection – has also been published in recent weeks:
Cox, M.A., Garwood, R.J., Behnsen, J., Hunt, J.N., Dalby, L.J., Martin, G.S., Maclaine, J.S., Wang, Z. & Cox, J.P. 2021. Olfactory flow in the sea catfish, Ariopsis felis (L.): Origin, regulation, and resampling. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology p.110933. doi: 10.1016/j.cbpa.2021.110933
17/12/2020
It's been a long since the last update of this site, in large part because of the COVID19 pandemic. This coincided with a relaunch of the degrees in the Department of Earth and Environmental Sciences. As a result I spent my time since June writing 1.5 course units for the first time, and then delivering them remotely. You can find the results at the two websites linked below:
The first of these sites (EART22101) has seven websites combining videos, quizzes, and other learning materials on topics surrounding evolution, and ancient life. The second (EART27201) has eight sites that focus on invertebrate macrofossils, and include 3D fossil models and the occasional virtual microscope. Materials are generally CC-BY and sources are always provided: they are available here in the hope that they are useful! There are other elements of course delivery provided through Blackboard (more quizzes, achievements to unlock) and via Zoom, which aren't included. Nevertheless, I hope they may be useful!
Early in lockdown, though, there was time for some science – and one of the results is a preprint which has just appeared on bioRxiv. You can find it by clicking on the link below:
In this paper, led by my colleague Nicolás Mongiardino Koch, we assess the impact of adding fossils to phylogenies using a simulation based approach, based on the Palaeoware package TREvoSim. We demonstrate that adding extinct species always improves the accuracy of a phylogenetic analysis, even in the presence of missing data. We also show that tip-dated phylogenies – i.e. those which simultaneously infer tree topology and divergence times – outperform all other methods of inference. You can find out more by reading the freely available preprint!
25/03/2020
A new paper, led by my colleague Jonathan Cox at the University of Bath, and a follow up to a paper released last year, has just appeared. This one uses CT scanning and CFD to study smell in the pike Esox lucius. As with the last such paper, although I am first author, this reflects authorship conventions in the field. I didn't lead the work, but did have the pleasure of conducting CT data collection. Details of this publication are as follows:
Garwood, R.J., Behnsen, J., Ramsey, A.T., Haysom, H.K., Dalby, L.J., Quilter, S.K., Maclaine, J.S., Wang, Z. & Cox, J.P.* 2020. The functional nasal anatomy of the pike, Esox lucius L. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 244: 110688. doi:10.1016/j.cbpa.2020.110688 (* = corresponding author)
06/03/2020
I am pleased to report that a new paper has just appeared, accompanied by two new Palaeoware evolutionary simulation software packages. The paper is:
Keating, J.N., Sansom, R.S.*, Sutton, M.D., Knight, C.G. & Garwood, R.J.* In press. Morphological phylogenetics evaluated using novel evolutionary simulations. Systematic Biology doi:10.1093/sysbio/syaa012 (* = corresponding author)
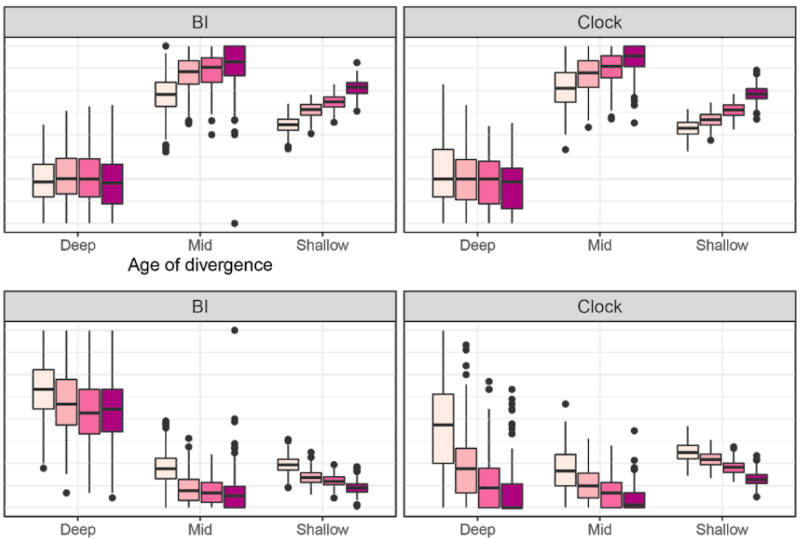
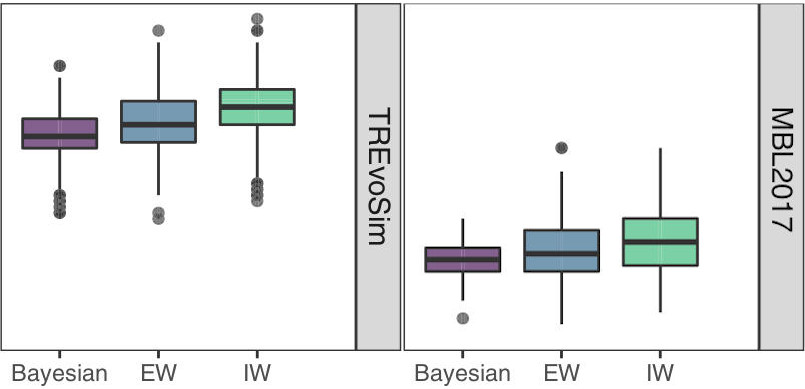
In it, my colleagues and I use two new evolutionary models to test methods of inferring evolutionary trees. We use the models to simultaneously generate trees and discrete character data, one in a stochastic manner, and one using an individual-based approach which includes selection. We use different approaches to infer trees from the character data, and then comparing how the approaches perform by measuring the distance between the true and inferred trees. There have been a number of recent works that have taken a simulation approach, generating stochastic data onto a set tree topology, which have generally suggested that Bayesian inference outperforms parsimony. Our study supports this idea, but also suggests this is because of the differences in search strategy and consensus methods used by each, rather than resulting from the optimality criterion they use. Our approach also allows us to demonstrate that stochastic- and selection-generated data behave differently, and that the amount of homoplasy in our datasets are not a good indicator of how methods of inference will perform. Rather morphological coherence (how strongly linked characters differences are to the evolutionary, or branch length distances between taxon paris) is a key consideration, and this is a bigger problem for selection-generated data. Morphology is thought to evolve through selection, and so we suggest that asking whether parsimony or Bayesian is better may not be the best approach for the field in future: both methods struggle with selection data. Rather if we can future research on modes of morphological evolution, and then use those to build models for probabilistic inference, we will do a better job of reconstructing trees in the future.
You can find releases of the two software packages/models we created for this paper by clicking the banner below. TREvoSim is our selection-based model. It uses a similar fitness-algorithm to REvoSim, but employs a different species concept, does not include sexual reproduction, eschews space, and has variable genome sizes. MBL2017 is a birth-death model, which simulates lineage splitting and extinction, as well as character evolution, in a stochastic manner. Both packages are available as binaries for Windows and Mac OS, and can be built on Linux systems using four lines of bash. We are releasing the two in the hope that they are versatile and useful tools for the community, and would very much welcome feedback, bug reports, and feature requests: these are best achieved using the issues system in GitHub, or by emailing us at palaeoware@gmail.com.